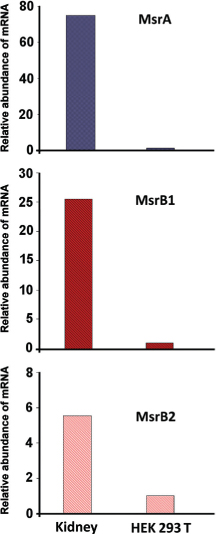
Figure 1. Levels of MsrA, MsrB1, and MsrB2 transcripts in human kidney and HEK 293T cells, analyzed by qRT-PCR. The mRNA levels of the three proteins were normalized over the levels of GAPDH mRNA.
ORIGINAL ARTICLE
Cesare Achilli, Annarita Ciana, Giampaolo Minetti
Laboratories of Biochemistry, Department of Biology and Biotechnology, University of Pavia, Pavia, Italy
The immortalized cell lines derived from human embryonic kidney, named HEK 293, are extensively used as models of human renal cells in in vitro studies. Nevertheless, ample evidence in the literature shows that HEK 293 cells display genotypic and phenotypic characteristics that differ substantially from primary kidney cells, with potential detrimental effects on the quality of the experimental results. Among the differences documented between HEK 293 and renal cells, there is an altered pattern of expression of many proteins involved in the development and physiological functions of the kidney. Methionine sulfoxide reductase (Msr) enzymes are ubiquitous components of the cellular machinery, evolved to counteract the damages inflicted to methionine residues by oxidative stress, particularly intense in kidney tissues. In this article, we have compared the levels of expression of several different Msr enzymes in human kidney and in a HEK 293 strain and have observed significant differences between the two cell types.
Keywords: HEK 293; kidney; methionine sulfoxide reductase; MsrA; MsrB
In 1951, the first immortalized human cell strain (designed as HeLa) was developed by propagation of poliomyelitis virus in tissue cultures prepared from epidermoid carcinoma of the cervix (1). Since then, more than 3600 new cell lines from over 150 species have been developed, revolutionizing the scientific world. In fact, immortalized cell lines are extensively used in many branches of basic biomedical research, from the study of gene function to generation of artificial tissues. They have several advantages over primary cells, such as the ability to proliferate virtually indefinitely, cost-effectiveness, easiness of handling, and no ethical restrictions associated with the use of animal or human tissues.
Cell culture studies provide a valuable complement for in vivo experiments; however, the necessary condition to use this strategy is that the immortalized cells display and maintain functional features as close as possible to related primary cells.
Among the many advantages there are, however, some critical issues that could dramatically affect the goodness of the results, such as contamination by other cell lines or by mycoplasma, and the misidentification of cell lines. About these problems, it is estimated that up to one third of cell lines are imposters (2–5).
Owing to their altered genome and karyotype, immortalized cell lines should be considered as cancerous cells, and consequently their native functions and responsiveness to stimuli could be different from those of related primary cells (6). In addition, the use of cultures at high-passage numbers could further induce the genotypic/phenotypic variability, which could also vary from one cell line to another (7). For these reasons, the immortalized cell lines cannot represent a priori a reliable model of their original source tissue and could provide controversial results. In order to avoid these mistakes, the current recommendations by the National Institutes of Health are to verify the results obtained using cell lines by running experiments with primary cells, and to authenticate the cell lines that are in use.
In 1977 it was reported, for the first time, that human embryonic kidney cells, obtained from an aborted fetus, were transformed by exposure to sheared human adenovirus type-5 DNA (8), and therefore they became an immortalized cell line. The superinfected cells, designated human embryonic kidney 293 (HEK 293) cells, and their derivative cell lines, such as the one transformed in 1987 by Epstein–Barr virus and named HEK 293T (9), are extensively used in biomedical and biotechnology research as a model of kidney epithelial cells. However, these renal cell strains express only a limited number of typical markers of the kidney development, such as those of the mesenchymal-epithelial transition, but not others, such as those of the differentiated tubular segments, proximal tubules, and collecting ducts. Surprisingly, HEK 293 cells express many neuromarkers, such as those typical of the neurofilament and other neurospecific metabolic enzymes (10).
HEK 293 cell line and its derivatives are among the most widely used human cell lines in biomedical experimentation, and the fact that properties of both neuron and kidney lineages coexist in the same strain could have significant implications for experiments in which a secure kidney cell control is required. In addition, it is unknown whether this cell line originally obtained from developing renal tissue can be representative of primary cells of an adult kidney, nor of which specific renal cell types. For these ambiguities, it would be safer not to use these cellular lines as a model of renal cells, especially for the study of kidney function.
The energy production necessary for the tubular reabsorption in the kidney relies on aerobic metabolism and oxidative phosphorylation, which implies, as a side effect, the release of high levels of reactive oxygen species (ROS). The renal cells activate several signaling pathways to maintain ROS at low levels. An excess of ROS would be very aggressive to several cellular components, including amino acids such as methionine, whose sulfur atom is easily oxidized to generate the two diastereoisomers of methionine sulfoxide. Since aberrant oxidation of methionine alters the physiological properties of many proteins, particularly high levels of methionine sulfoxide reductase enzymes (Msr) are expressed, among the other tissues, in the kidney. Msrs are ubiquitous enzymes of the cellular weaponry that has evolved to prevent or revert the damage inflicted by elevated levels of oxidants to sensitive cellular components. Msrs, in particular, perform the reconversion of methionine sulfoxide to methionine (11–13). Msrs were classified on the basis of their stereoselectivity toward the two diastereoisomers of methionine sulfoxide: MsrAs reduce methionine-S-sulfoxide, whereas MsrBs reduce methionine-R-sulfoxide (14).
Very little is known on the expression of Msr enzymes in the cell lineages used in vitro as a model of human kidney cells. Considering the importance of this class of enzymes in the protection of the kidney against oxidative stress, and the extensive and inescapable use of immortalized cell lines for the in vitro studies on renal cell function, this lacuna should be filled up. Here, we have therefore compared the levels of some Msr enzymes in human kidney cells and HEK 293T lineage by quantitative analysis of their transcripts.
HEK 293T cells were grown in adhesion in Dulbecco’s modified Eagle’s medium (supplemented with 10% of heat-inactivated bovine fetal serum, L-glutamine, streptomycin, and penicillin) at 37°C in the presence of 5% CO2 and harvested at about 90% confluence by gentle stirring. The cells were then washed once with PBS, centrifuged at 500 g for 10 min at 4°C, and stored at −80°C.
The human kidney cDNA required to perform the quantitative reverse transcription polymerase chain reaction (qRT-PCR) was purchased from Thermo Fisher Scientific (Milano, Italy), while RNA of HEK 293T was extracted and then reverse transcribed, as follows: Total RNA was extracted using Trizol® reagent (Thermo Fisher Scientific, Milano, Italy) according to the supplier’s instructions. The RNA obtained was dissolved in diethyl pyrocarbonate (DEPC)-treated MilliQ® water and the concentration was determined by spectrophotometric assay at 260 nm. After extraction, RNA was immediately reverse transcribed. First-strand cDNA was synthesized using 2–10 μg total RNA using the High Capacity cDNA Archive Kit (Thermo Fisher Scientific, Milano, Italy) according to the manufacturer’s recommendations. The samples were then stored at −80°C.
The expression levels of MsrA, MsrB1, MsrB2, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were evaluated using qRT-PCR based on the TaqMan® Gene Expression Assay (Thermo Fisher Scientific, Milano, Italy; Assay ID: Hs00737165_m1 for MsrA; Hs00249482_m1 for MsrB1; Hs00255292_m1 for MsrB2; and Hs99999905_m1 for GAPDH) according to the manufacturer’s instruction. Standard curves were constructed by serial dilution of cDNA samples for the different Msrs with DEPC-treated MilliQ® water. The amplification reactions were performed in a 96-well microtiter plate in a thermocycler for quantitative PCR (Mx 3000P and MxPro software, Agilent Technologies, Milano, Italy). The normalization of the results was carried out by comparison with the relative expression levels of the housekeeping gene GAPDH in the same cell type. Through the calibration curve the relative abundance of the transcript was obtained for each sample. The value obtained for each Msr gene in every cell type was compared with the corresponding value obtained for GAPDH. This procedure allowed a direct comparison of the expression levels of the transcripts in the examined cell types.
In this study, we examined the mRNA levels of MsrA, MsrB1, and MsrB2, the typical forms of Msrs expressed in mammalian tissues, and for which the presence in the human renal tissues has been documented for adult and embryonic kidney (11–13). The presence of MsrA, MsrB1, and MsrB2 transcripts was evaluated by qRT-PCR in cDNA obtained from HEK 293T cells and total human kidney. The latter, which ideally includes the cDNA obtained from all the cell types of which the adult kidney is composed, represents an excellent reference to perform a comparison with a HEK 293 lineage, whose exact attribution is still ignored and, paradoxically, should be a heterogeneous mix of almost all the cell types present in the body. This technique allows the quantification of transcripts for a gene of interest by comparing them with the levels of transcript of a housekeeping gene in the same cell type, thus permitting direct comparison of absolute levels of transcripts in different tissues. The results obtained were normalized over the expression levels of GAPDH, used as housekeeping gene. Using this method it could be verified that the levels of MsrA, MsrB1, and MsrB2 transcripts were, respectively, 80, 25, and 6 times lower in HEK 293T than in the human kidney tissue (Figure 1).

Figure 1. Levels of MsrA, MsrB1, and MsrB2 transcripts in human kidney and HEK 293T cells, analyzed by qRT-PCR. The mRNA levels of the three proteins were normalized over the levels of GAPDH mRNA.
The debate on the authenticity of HEK 293 cells has been going on for 20 years and does not seem to have reached a conclusion. Every study conducted to unmask in an incontrovertible manner the “double face” of this immortalized cell line represents a small step toward the truth. The data collected in this paper seem to corroborate the evidence widely described in the literature concerning the inadequacy of HEK 293 lineage as a model of renal cells.
The results are also in agreement with enzymatic assays and Western blotting analysis preliminarily conducted on cell extracts of HEK 293T, essentially as we reported for several human blood cells and murine organs (13), in which very low levels of Msr activity and protein expression emerged in this cell line (data not shown).
The low expression of Msrs in HEK 293T cells could be due to the alteration of the energy metabolism that is commonly found in tumor cells, with enhanced glycolytic activity and diminished mitochondrial metabolic capacity (15). A slow mitochondrial cellular respiration is associated with a decreased production of ROS; therefore, tumor cells could also thrive in the absence of elaborate enzymatic defense/repair systems.
The limited expression of Msrs is also in agreement with the hypothesis that HEK 293T cells are not actually renal cells but more likely a neuronal lineage (16). In neurons, lower levels of expression of Msrs with respect to kidney have been described (11, 12). Data on the neuronal genotype of HEK 293T, and the lower expression of Msr enzymes in most human brain tissues compared to renal cells, seem to be consistent with the results presented here. Our observations seem to corroborate the data reported in the literature, which show how the HEK 293 cell line and its derivatives, including HEK 293T, are not cell lines suitable as a model of renal cells, and therefore should be used with caution in studies focused on the physiology of kidney cells (10).
HEK 293 cells and their derivative HEK 293T are defined as human embryonic kidney cells and under this denomination are widely used experimentally in cell biology studies. The long-going debate around their true origin, which is probably from adrenal cells that display many features of neuronal cells and could have been preferentially transformed by adenovirus when the cell line was originally established, could not help to officially change their denomination. We have reported here that HEK 293T cells express very low levels of Msr enzymes (MsrA, MsrB1, and MsrB2), which instead are highly expressed in human kidney cells. This data coincides with the hypothesis, suggested by several researchers, that HEK 293T cells actually are not renal cells and, for this reason, are not a good model of human kidney cells.
Part of this work was supported with funds from the “Ministero dell’Istruzione, Università e Ricerca Scientifica” Italy (PRIN 2006).
The author declares no conflicts of interest with respect to research, authorship, and/or publication of this article.